Utah Census Data: Measles Outbreak Modeling Comparison
utah_census_comparison.Rmd
library(multigroup.vaccine)
library(socialmixr)
# Use the included example data file to avoid download issues during package building
census_csv <- getCensusDataPath()Introduction
This vignette demonstrates how to use the
getCensusData() function to retrieve real U.S. Census
Bureau population data for Utah counties and use it in measles outbreak
modeling. We’ll compare how different counties with varying demographic
structures respond to measles outbreaks under different vaccination
scenarios.
Note: This vignette uses the included example census data file to ensure it works offline and during package building.
Measles Model Setup
For measles outbreak modeling, we’ll use the standard age groups from the measles age-structured model:
Getting Census Data for Utah Counties
Let’s retrieve population data for three diverse Utah counties:
utah_fips <- getStateFIPS("Utah")
# Get data for three counties with different characteristics
counties <- c("Salt Lake County", "Utah County", "Washington County")
county_data_list <- list()
for (county in counties) {
data <- getCensusData(
state_fips = utah_fips,
county_name = county,
year = 2024,
age_groups = agelims,
csv_path = census_csv
)
county_data_list[[county]] <- data
cat("\n", county, ":\n", sep = "")
cat(" Total population:", format(data$total_pop, big.mark = ","), "\n")
cat(" Age distribution:\n")
for (i in seq_along(data$age_labels)) {
pct <- 100 * data$age_pops[i] / data$total_pop
cat(sprintf(" %s: %s (%.1f%%)\n",
data$age_labels[i],
format(data$age_pops[i], big.mark = ","),
pct))
}
}
#> Reading census data from: /home/runner/work/_temp/Library/multigroup.vaccine/extdata/cc-est2024-syasex-49.csv
#> Aggregating ages 0 to 0: sum = 14732
#> Aggregating ages 1 to 4: sum = 57711
#> Aggregating ages 5 to 11: sum = 111884
#> Aggregating ages 12 to 17: sum = 108100
#> Aggregating ages 18 to 24: sum = 125472
#> Aggregating ages 25 to 44: sum = 379015
#> Aggregating ages 45 to 69: sum = 318909
#>
#> Salt Lake County:
#> Total population: 1,216,274
#> Age distribution:
#> under1: 14,732 (1.2%)
#> 1to4: 57,711 (4.7%)
#> 5to11: 111,884 (9.2%)
#> 12to17: 108,100 (8.9%)
#> 18to24: 125,472 (10.3%)
#> 25to44: 379,015 (31.2%)
#> 45to69: 318,909 (26.2%)
#> 70plus: 100,451 (8.3%)
#> Reading census data from: /home/runner/work/_temp/Library/multigroup.vaccine/extdata/cc-est2024-syasex-49.csv
#> Aggregating ages 0 to 0: sum = 12053
#> Aggregating ages 1 to 4: sum = 48596
#> Aggregating ages 5 to 11: sum = 88402
#> Aggregating ages 12 to 17: sum = 78366
#> Aggregating ages 18 to 24: sum = 125580
#> Aggregating ages 25 to 44: sum = 210551
#> Aggregating ages 45 to 69: sum = 143596
#>
#> Utah County:
#> Total population: 747,234
#> Age distribution:
#> under1: 12,053 (1.6%)
#> 1to4: 48,596 (6.5%)
#> 5to11: 88,402 (11.8%)
#> 12to17: 78,366 (10.5%)
#> 18to24: 125,580 (16.8%)
#> 25to44: 210,551 (28.2%)
#> 45to69: 143,596 (19.2%)
#> 70plus: 40,090 (5.4%)
#> Reading census data from: /home/runner/work/_temp/Library/multigroup.vaccine/extdata/cc-est2024-syasex-49.csv
#> Aggregating ages 0 to 0: sum = 2263
#> Aggregating ages 1 to 4: sum = 9687
#> Aggregating ages 5 to 11: sum = 18808
#> Aggregating ages 12 to 17: sum = 18446
#> Aggregating ages 18 to 24: sum = 19948
#> Aggregating ages 25 to 44: sum = 49399
#> Aggregating ages 45 to 69: sum = 55271
#>
#> Washington County:
#> Total population: 207,943
#> Age distribution:
#> under1: 2,263 (1.1%)
#> 1to4: 9,687 (4.7%)
#> 5to11: 18,808 (9.0%)
#> 12to17: 18,446 (8.9%)
#> 18to24: 19,948 (9.6%)
#> 25to44: 49,399 (23.8%)
#> 45to69: 55,271 (26.6%)
#> 70plus: 34,121 (16.4%)Scenario 1: Current Vaccination Coverage
Let’s model outbreaks under current estimated vaccination coverage levels in Utah.
# Based on school data and estimates
current_coverage <- c(0, 0.89, 0.949, 0.950, 0.95, 0.95, 0.95, 1)Salt Lake County - Current Coverage
slc_data <- county_data_list[["Salt Lake County"]]
slc_current <- multigroup.vaccine:::getOutputTable(
agelims = agelims,
agepops = slc_data$age_pops,
agecovr = current_coverage,
ageveff = ageveff,
initgrp = initgrp
)
cat("Salt Lake County - Current Vaccination Coverage\n")
#> Salt Lake County - Current Vaccination Coverage
print(as.data.frame(slc_current), row.names = FALSE)
#> R0 R0local Rv pEscape escapeInfTot under1 1to4 5to11 12to17 18to24
#> 10 11.42435 0.9402893 0.000 0 0 0 0 0 0
#> 11 12.56679 1.0343182 0.075 4922 400 448 599 846 508
#> 12 13.70922 1.1283471 0.093 17506 1530 1654 2070 2638 1812
#> 13 14.85166 1.2223761 0.146 28583 2644 2760 3275 3908 2954
#> 14 15.99409 1.3164050 0.239 38133 3694 3727 4241 4838 3926
#> 15 17.13653 1.4104339 0.272 46321 4661 4559 5015 5539 4748
#> 16 18.27896 1.5044628 0.287 53345 5544 5269 5639 6078 5440
#> 17 19.42140 1.5984918 0.347 59386 6345 5875 6146 6501 6026
#> 18 20.56383 1.6925207 0.380 64604 7069 6392 6560 6837 6522
#> 25to44 45to69 70+
#> 0 0 0
#> 1375 746 0
#> 5014 2788 0
#> 8306 4736 0
#> 11175 6530 0
#> 13640 8158 0
#> 15747 9627 0
#> 17546 10948 0
#> 19086 12137 0Utah County - Current Coverage
utah_data <- county_data_list[["Utah County"]]
utah_current <- multigroup.vaccine:::getOutputTable(
agelims = agelims,
agepops = utah_data$age_pops,
agecovr = current_coverage,
ageveff = ageveff,
initgrp = initgrp
)
cat("\nUtah County - Current Vaccination Coverage\n")
#>
#> Utah County - Current Vaccination Coverage
print(as.data.frame(utah_current), row.names = FALSE)
#> R0 R0local Rv pEscape escapeInfTot under1 1to4 5to11 12to17 18to24
#> 10 13.36528 1.106482 0.101 9582 983 1126 1361 1603 1614
#> 11 14.70180 1.217131 0.129 18227 2025 2219 2503 2724 3042
#> 12 16.03833 1.327779 0.206 25527 3016 3168 3391 3507 4210
#> 13 17.37486 1.438427 0.251 31653 3929 3970 4080 4075 5157
#> 14 18.71139 1.549075 0.276 36800 4754 4640 4618 4499 5924
#> 15 20.04792 1.659724 0.348 41143 5495 5198 5043 4821 6549
#> 16 21.38444 1.770372 0.355 44826 6156 5663 5381 5071 7060
#> 17 22.72097 1.881020 0.394 47967 6745 6051 5654 5267 7482
#> 18 24.05750 1.991668 0.406 50660 7269 6378 5875 5423 7831
#> 25to44 45to69 70+
#> 2025 870 0
#> 3967 1747 0
#> 5671 2563 0
#> 7136 3306 0
#> 8386 3978 0
#> 9452 4585 0
#> 10362 5133 0
#> 11140 5628 0
#> 11808 6075 0Washington County - Current Coverage
wash_data <- county_data_list[["Washington County"]]
wash_current <- multigroup.vaccine:::getOutputTable(
agelims = agelims,
agepops = wash_data$age_pops,
agecovr = current_coverage,
ageveff = ageveff,
initgrp = initgrp
)
cat("\nWashington County - Current Vaccination Coverage\n")
#>
#> Washington County - Current Vaccination Coverage
print(as.data.frame(wash_current), row.names = FALSE)
#> R0 R0local Rv pEscape escapeInfTot under1 1to4 5to11 12to17 18to24
#> 10 10.86329 0.8823416 0.000 0 0 0 0 0 0
#> 11 11.94962 0.9705758 0.000 0 0 0 0 0 0
#> 12 13.03595 1.0588100 0.079 1130 86 109 153 231 117
#> 13 14.12227 1.1470441 0.112 2737 225 276 363 493 285
#> 14 15.20860 1.2352783 0.140 4217 369 437 545 687 441
#> 15 16.29493 1.3235125 0.204 5541 509 585 696 834 580
#> 16 17.38126 1.4117466 0.255 6711 642 716 820 946 701
#> 17 18.46759 1.4999808 0.268 7741 767 831 922 1035 806
#> 18 19.55392 1.5882150 0.322 8647 882 931 1006 1104 898
#> 25to44 45to69 70+
#> 0 0 0
#> 0 0 0
#> 250 184 0
#> 625 469 0
#> 984 754 0
#> 1312 1026 0
#> 1605 1281 0
#> 1865 1516 0
#> 2092 1733 0Visualization: Comparing Counties
Let’s visualize the outbreak potential across counties:
# Extract R0=15 results for comparison
r0_15_idx <- which(slc_current[, "R0"] == 15)
counties_names <- c("Salt Lake", "Utah", "Washington")
escape_totals <- c(
slc_current[r0_15_idx, "escapeInfTot"],
utah_current[r0_15_idx, "escapeInfTot"],
wash_current[r0_15_idx, "escapeInfTot"]
)
total_pops <- c(
slc_data$total_pop,
utah_data$total_pop,
wash_data$total_pop
)
escape_pcts <- 100 * escape_totals / total_pops
par(mfrow = c(1, 2), mar = c(5, 4, 4, 2))
# Absolute numbers
barplot(escape_totals,
names.arg = counties_names,
main = "Potential Outbreak Size (R0=15)\nCurrent Coverage",
ylab = "Total Infections",
col = "coral",
las = 2)
# Percentages
barplot(escape_pcts,
names.arg = counties_names,
main = "% of Population (R0=15)\nCurrent Coverage",
ylab = "% of Population Infected",
col = "steelblue",
las = 2)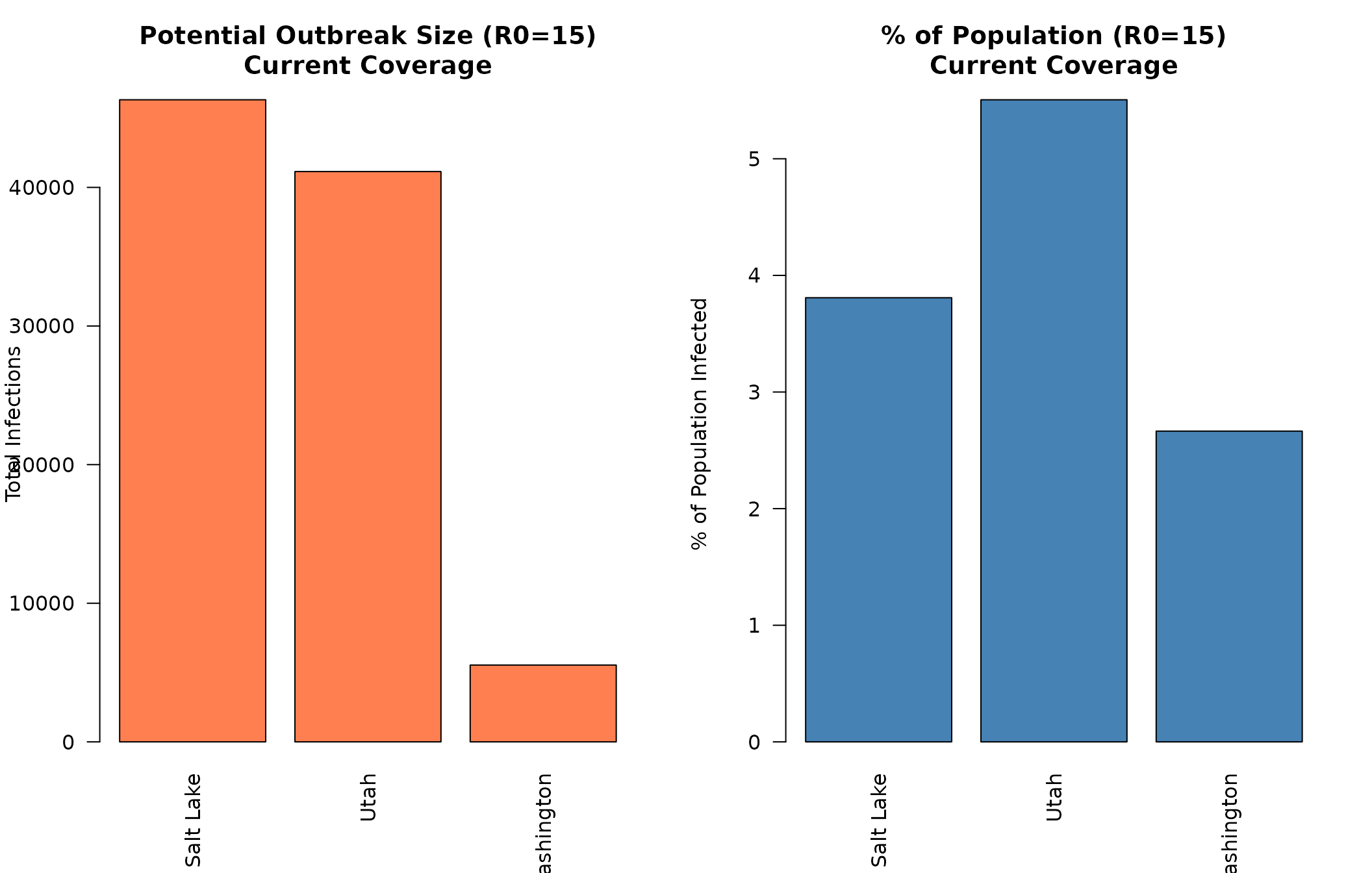
Scenario 2: Reduced Vaccination Coverage
What happens if vaccination coverage drops by 10% across all age groups?
reduced_coverage <- current_coverage * 0.9 # 10% reduction
reduced_coverage[1] <- 0 # Keep under-1 at 0
slc_reduced <- multigroup.vaccine:::getOutputTable(
agelims = agelims,
agepops = slc_data$age_pops,
agecovr = reduced_coverage,
ageveff = ageveff,
initgrp = initgrp
)
utah_reduced <- multigroup.vaccine:::getOutputTable(
agelims = agelims,
agepops = utah_data$age_pops,
agecovr = reduced_coverage,
ageveff = ageveff,
initgrp = initgrp
)
wash_reduced <- multigroup.vaccine:::getOutputTable(
agelims = agelims,
agepops = wash_data$age_pops,
agecovr = reduced_coverage,
ageveff = ageveff,
initgrp = initgrp
)
cat("Salt Lake County - Reduced Coverage (-10%)\n")
#> Salt Lake County - Reduced Coverage (-10%)
print(as.data.frame(slc_reduced), row.names = FALSE)
#> R0 R0local Rv pEscape escapeInfTot under1 1to4 5to11 12to17 18to24
#> 10 11.42435 1.979104 0.455 149540 7095 10043 15806 16378 16556
#> 11 12.56679 2.177015 0.487 161833 8031 10966 16683 16985 17699
#> 12 13.70922 2.374925 0.535 171440 8836 11687 17311 17399 18545
#> 13 14.85166 2.572836 0.569 179053 9532 12255 17768 17688 19180
#> 14 15.99409 2.770746 0.608 185157 10135 12706 18105 17892 19662
#> 15 17.13653 2.968657 0.634 190105 10660 13067 18357 18038 20032
#> 16 18.27896 3.166567 0.663 194153 11118 13358 18547 18144 20319
#> 17 19.42140 3.364477 0.682 197493 11519 13594 18691 18222 20542
#> 18 20.56383 3.562388 0.689 200269 11871 13787 18801 18279 20717
#> 25to44 45to69 70+
#> 47891 32417 3355
#> 51657 35938 3873
#> 54497 38823 4342
#> 56661 41202 4767
#> 58327 43176 5154
#> 59620 44824 5507
#> 60632 46206 5829
#> 61428 47372 6125
#> 62059 48358 6397Impact of Reduced Coverage
# Compare current vs reduced for R0=15
escape_reduced <- c(
slc_reduced[r0_15_idx, "escapeInfTot"],
utah_reduced[r0_15_idx, "escapeInfTot"],
wash_reduced[r0_15_idx, "escapeInfTot"]
)
comparison_matrix <- rbind(escape_totals, escape_reduced)
colnames(comparison_matrix) <- counties_names
rownames(comparison_matrix) <- c("Current", "Reduced -10%")
barplot(comparison_matrix,
beside = TRUE,
main = "Impact of 10% Reduction in Vaccination Coverage\n(R0=15)",
xlab = "County",
ylab = "Total Infections",
col = c("steelblue", "coral"),
legend.text = rownames(comparison_matrix),
args.legend = list(x = "topleft", bty = "n"))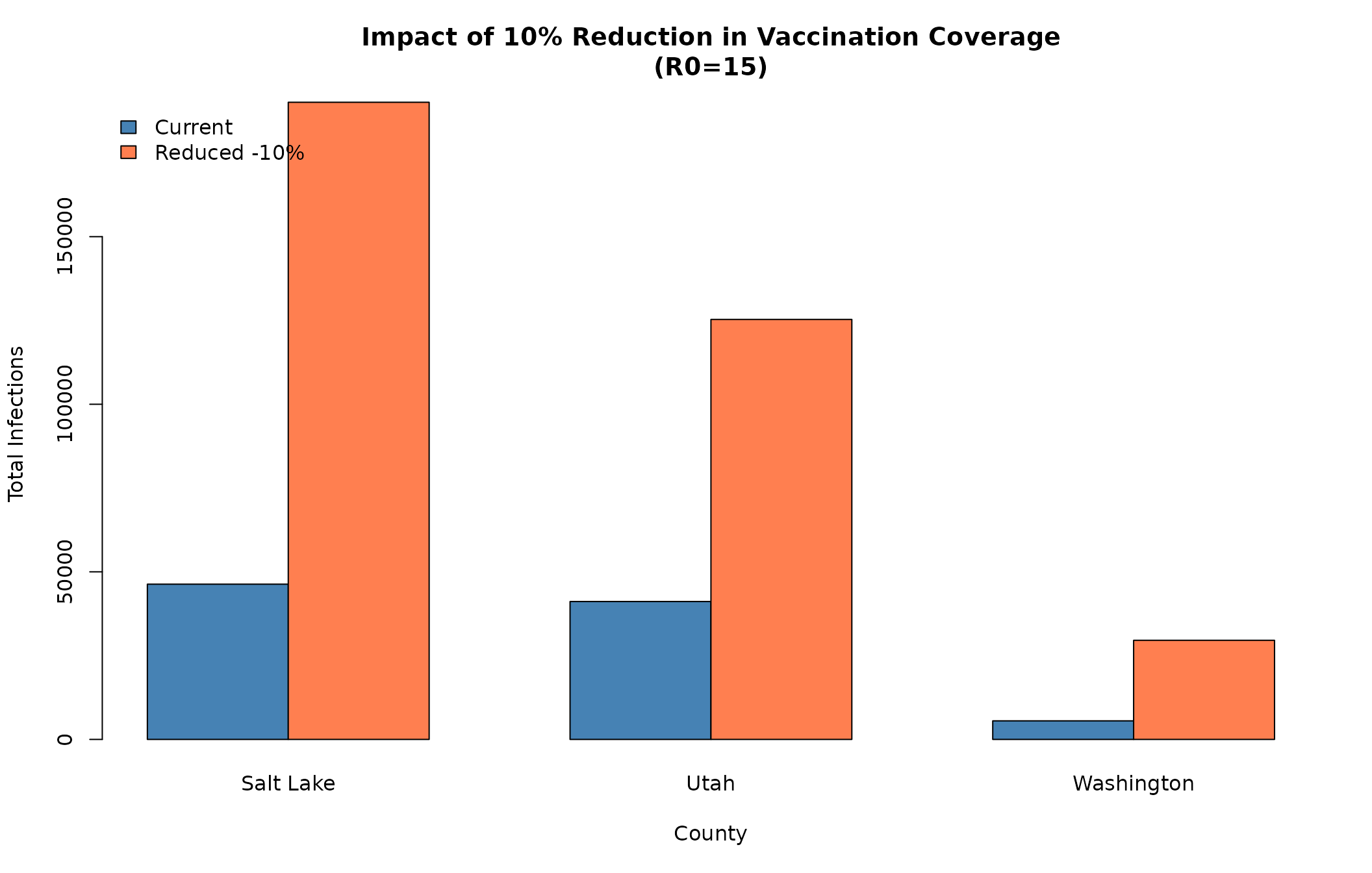
# Calculate percent increase
pct_increase <- 100 * (escape_reduced - escape_totals) / escape_totals
cat("\nPercent increase in outbreak size with 10% coverage reduction:\n")
#>
#> Percent increase in outbreak size with 10% coverage reduction:
for (i in seq_along(counties_names)) {
cat(sprintf(" %s: +%.1f%%\n", counties_names[i], pct_increase[i]))
}
#> Salt Lake: +310.4%
#> Utah: +204.6%
#> Washington: +433.7%Scenario 3: Age-Specific Vaccination Gaps
What if vaccination coverage is particularly low in school-age children (5-17)?
school_gap_coverage <- current_coverage
school_gap_coverage[3] <- 0.75 # 5-11: reduced from 85.2% to 75%
school_gap_coverage[4] <- 0.75 # 12-17: reduced from 87.9% to 75%
slc_schoolgap <- multigroup.vaccine:::getOutputTable(
agelims = agelims,
agepops = slc_data$age_pops,
agecovr = school_gap_coverage,
ageveff = ageveff,
initgrp = initgrp
)
utah_schoolgap <- multigroup.vaccine:::getOutputTable(
agelims = agelims,
agepops = utah_data$age_pops,
agecovr = school_gap_coverage,
ageveff = ageveff,
initgrp = initgrp
)
wash_schoolgap <- multigroup.vaccine:::getOutputTable(
agelims = agelims,
agepops = wash_data$age_pops,
agecovr = school_gap_coverage,
ageveff = ageveff,
initgrp = initgrp
)
cat("Salt Lake County - School-Age Vaccination Reduction\n")
#> Salt Lake County - School-Age Vaccination Reduction
print(as.data.frame(slc_schoolgap), row.names = FALSE)
#> R0 R0local Rv pEscape escapeInfTot under1 1to4 5to11 12to17 18to24
#> 10 11.42435 2.612491 0.344 95041 4956 5198 26503 27594 5429
#> 11 12.56679 2.873740 0.371 101832 5740 5820 27564 28171 6022
#> 12 13.70922 3.134989 0.411 107627 6480 6367 28326 28561 6540
#> 13 14.85166 3.396238 0.452 112603 7173 6843 28880 28829 6990
#> 14 15.99409 3.657487 0.487 116895 7816 7257 29286 29015 7379
#> 15 17.13653 3.918736 0.482 120612 8409 7614 29585 29144 7715
#> 16 18.27896 4.179985 0.569 123841 8954 7923 29807 29235 8003
#> 17 19.42140 4.441234 0.568 126656 9453 8190 29973 29299 8252
#> 18 20.56383 4.702483 0.600 129117 9909 8420 30097 29344 8465
#> 25to44 45to69 70+
#> 15880 9481 0
#> 17706 10810 0
#> 19308 12044 0
#> 20705 13182 0
#> 21917 14226 0
#> 22966 15179 0
#> 23871 16048 0
#> 24653 16838 0
#> 25326 17556 0Age-Specific Impact
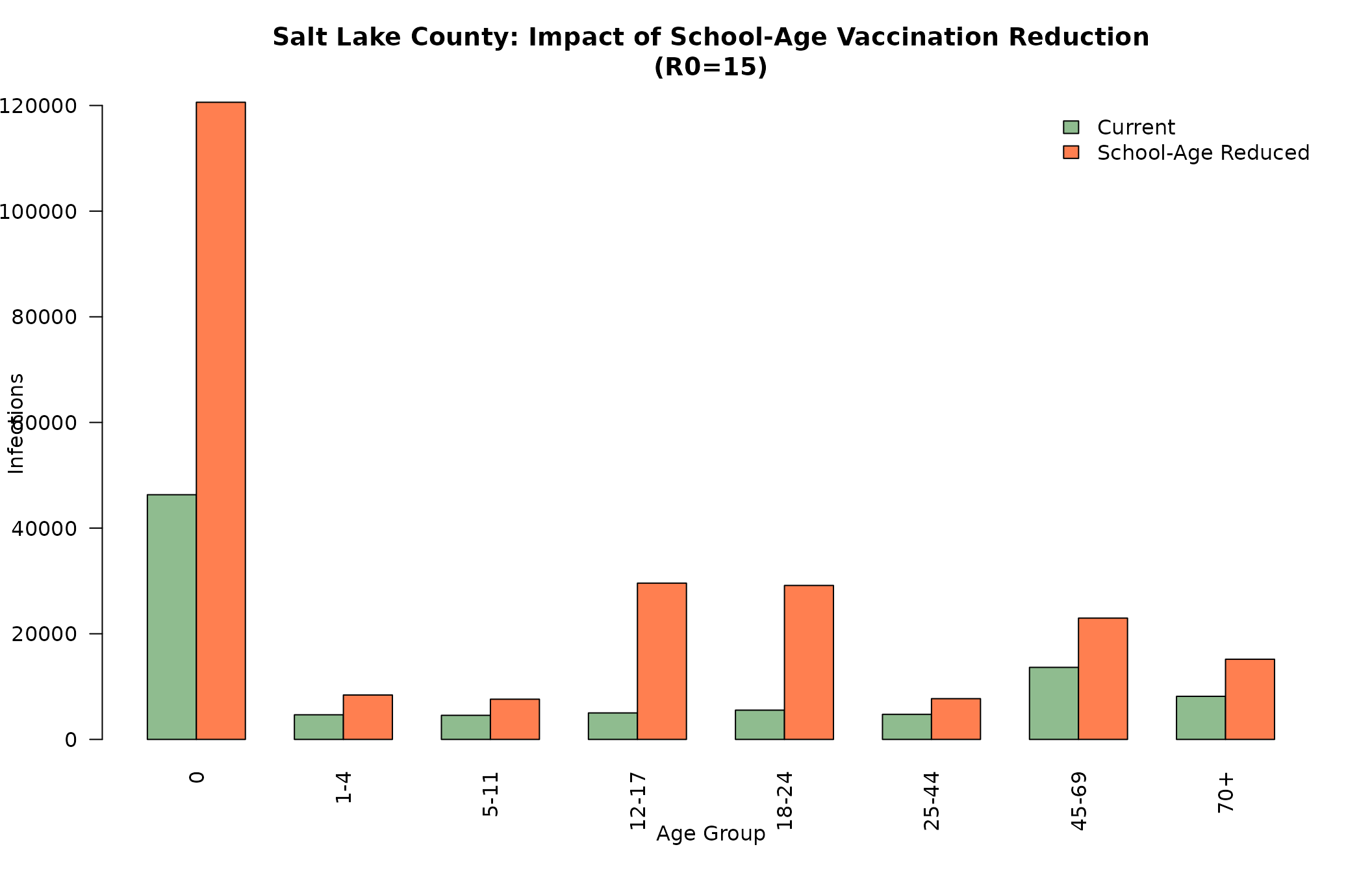
# Compare infections by age group for Salt Lake County
age_labels_short <- c("0", "1-4", "5-11", "12-17", "18-24", "25-44", "45-69", "70+")
# Get age-specific infections for R0=15
slc_current_ages <- as.numeric(slc_current[r0_15_idx, 5:12])
slc_schoolgap_ages <- as.numeric(slc_schoolgap[r0_15_idx, 5:12])
age_comparison <- rbind(slc_current_ages, slc_schoolgap_ages)
colnames(age_comparison) <- age_labels_short
rownames(age_comparison) <- c("Current", "School-Age Reduced")
barplot(age_comparison,
beside = TRUE,
main = "Salt Lake County: Impact of School-Age Vaccination Reduction\n(R0=15)",
xlab = "Age Group",
ylab = "Infections",
col = c("darkseagreen", "coral"),
legend.text = rownames(age_comparison),
args.legend = list(x = "topright", bty = "n"),
las = 2)