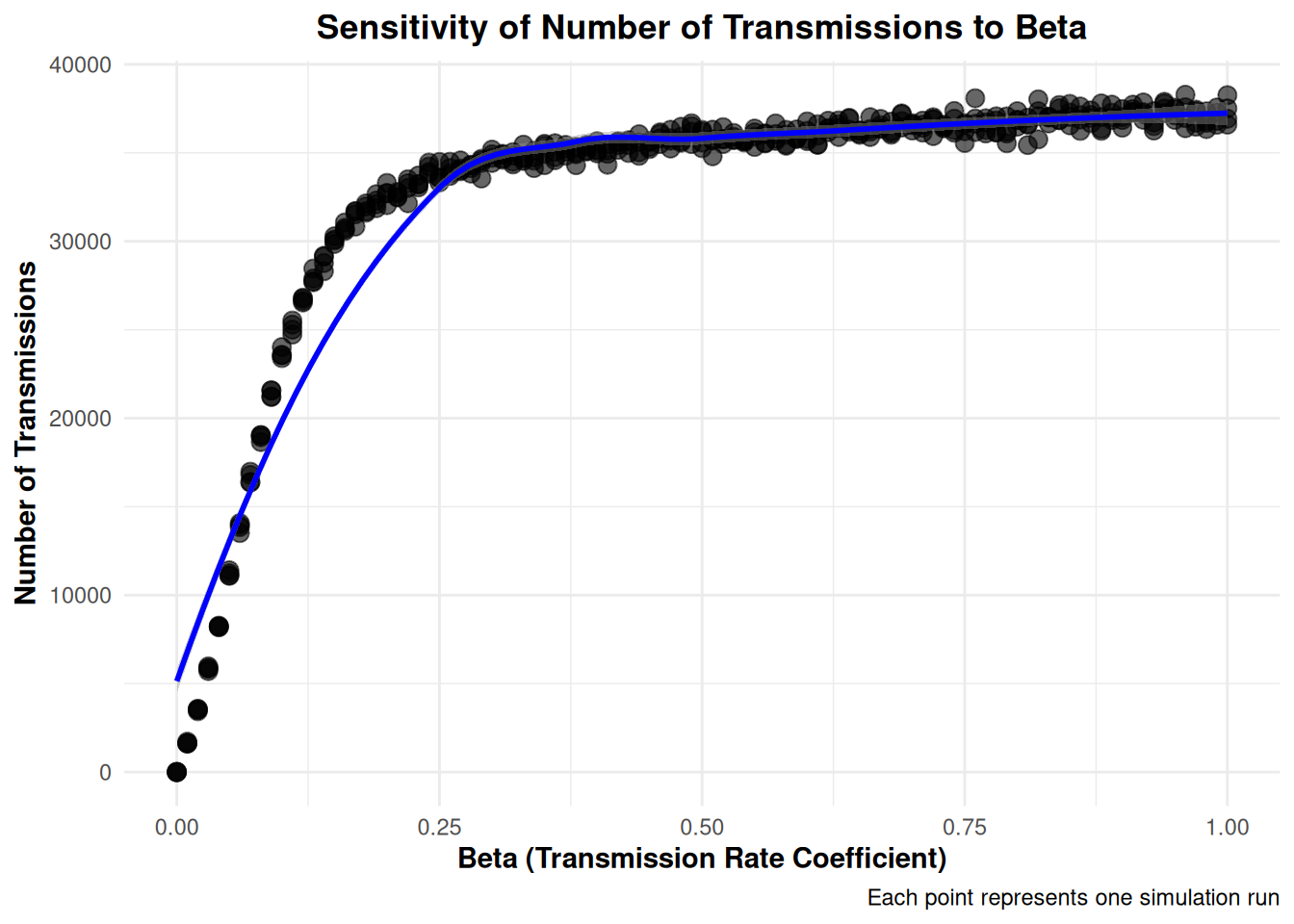
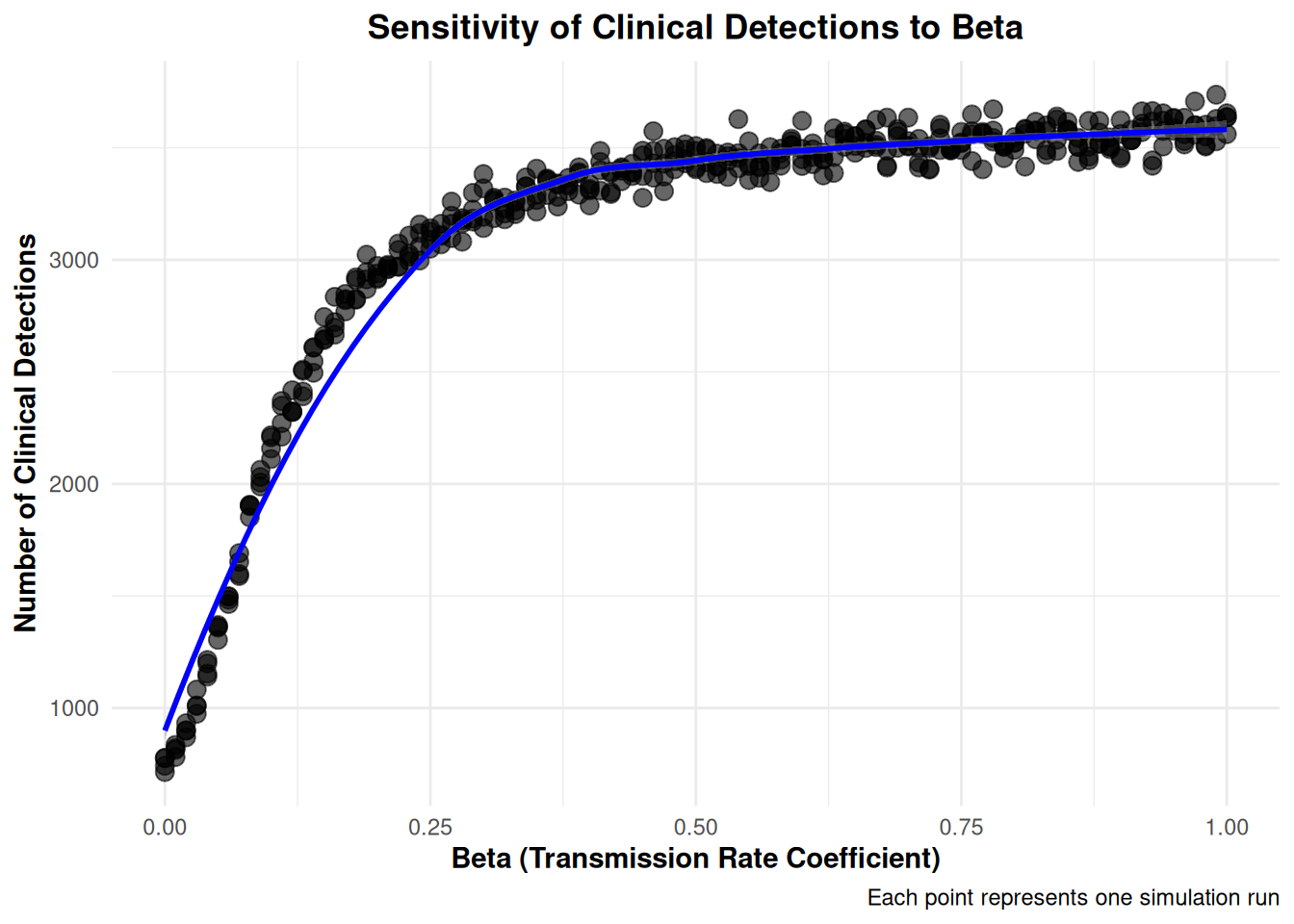
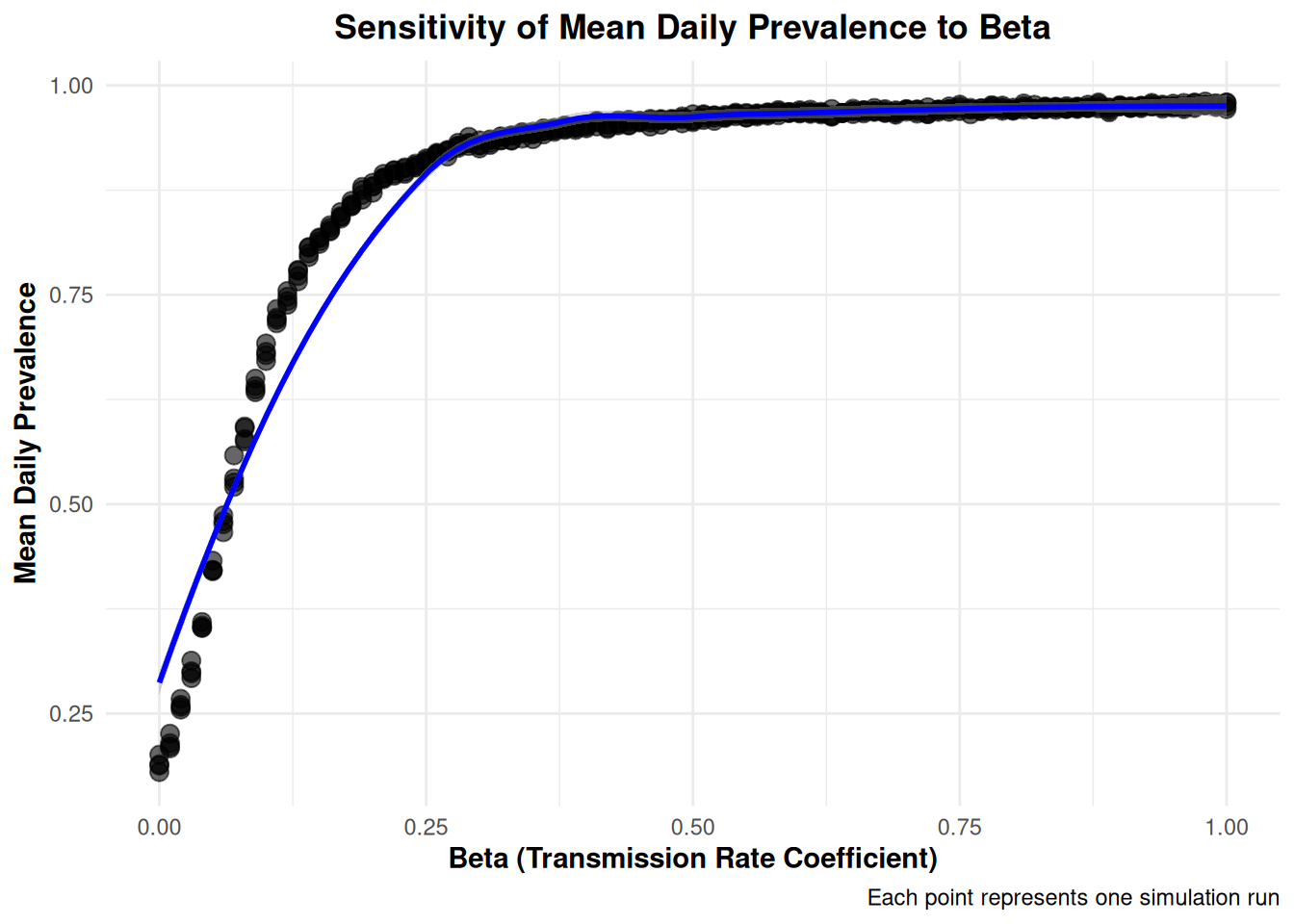
Rows: 404
Columns: 34
$ run <dbl> 1, 19, 37, 55, 73, 91, 109, 127, 145, …
$ DoActiveSurveillance <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
$ IsolationEffectiveness <dbl> 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5…
$ DaysBetweenTests <dbl> 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7,…
$ tick <dbl> 5474, 5474, 5474, 5474, 5474, 5474, 54…
$ ClinicalDetections <dbl> 777, 2922, 3358, 3408, 3498, 3623, 158…
$ MeanDailyPrevalence <dbl> 0.1801766, 0.8621268, 0.9486873, 0.965…
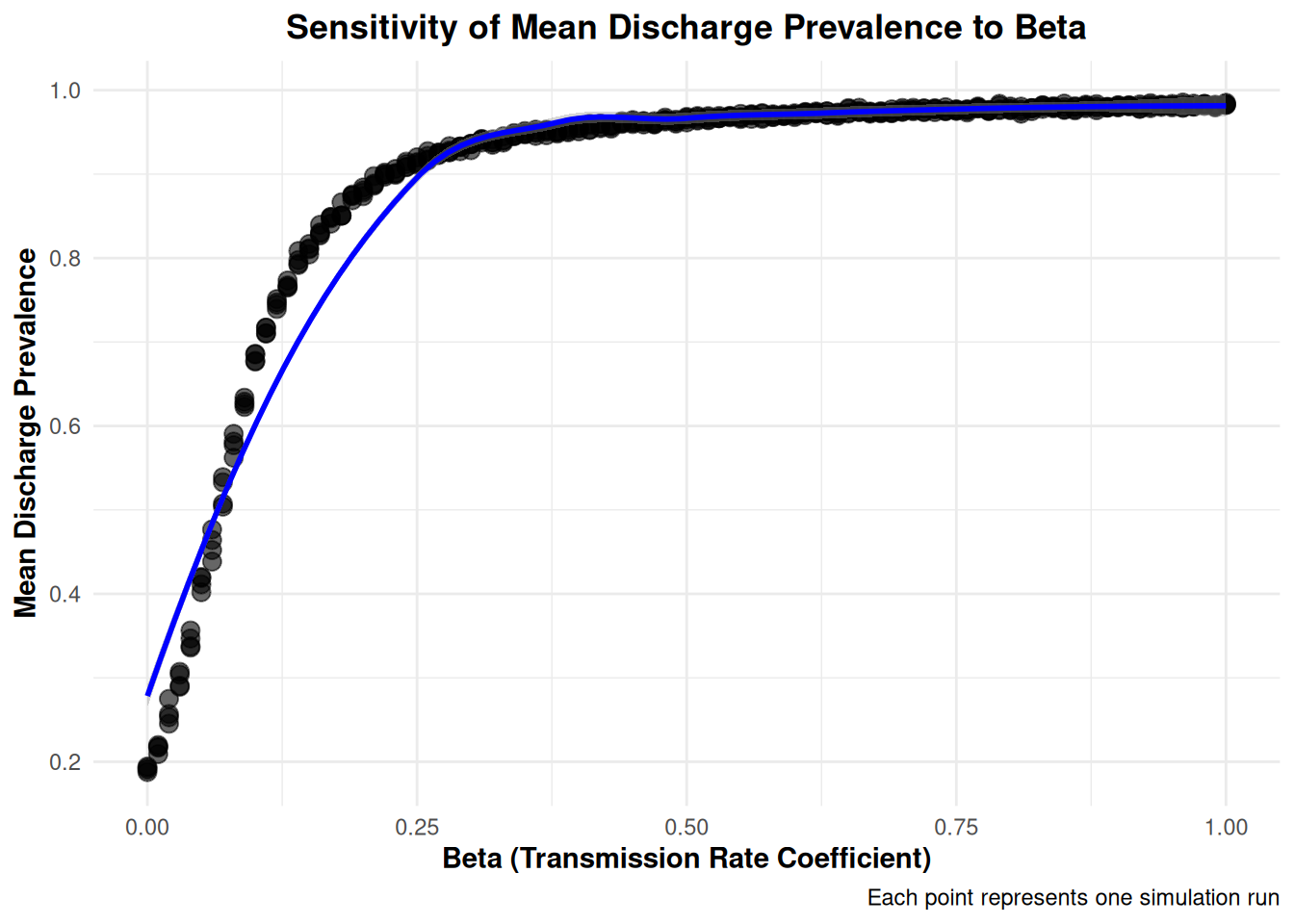
$ MeanDischargePrevalence <dbl> 0.1943099, 0.8664326, 0.9491032, 0.969…
$ ImportationPrevalence <dbl> 0.2090799, 0.2095499, 0.2051085, 0.210…
$ NumberOfTransmissions <dbl> 0, 32138, 34586, 35672, 36904, 36422, …
$ allowImportationsDuringBurnIn <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
$ isBatchRun <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
$ isolatePatientWhenDetected <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TR…
$ doActiveSurveillanceAfterBurnIn <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FAL…
$ isolationEffectiveness <dbl> 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5…
$ diseaseName <chr> "CRE", "CRE", "CRE", "CRE", "CRE", "CR…
$ shape1 <dbl> 7.60197, 7.60197, 7.60197, 7.60197, 7.…
$ shape2 <dbl> 1.23279, 1.23279, 1.23279, 1.23279, 1.…
$ importationRate <dbl> 0.206, 0.206, 0.206, 0.206, 0.206, 0.2…
$ isActiveSurveillanceAgent <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TR…
$ beta <dbl> 0.00, 0.18, 0.36, 0.54, 0.72, 0.90, 0.…
$ extraIteration <dbl> 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 3,…
$ prob1 <dbl> 0.62531, 0.62531, 0.62531, 0.62531, 0.…
$ midstaySurveillanceAdherence <dbl> 0.954, 0.954, 0.954, 0.954, 0.954, 0.9…
$ daysBetweenTests <dbl> 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7,…
$ simulationDurationAfterBurnIn <dbl> 5475, 5475, 5475, 5475, 5475, 5475, 54…
$ randomSeed <dbl> -929615374, -929609984, -929571943, -9…
$ admissionSurveillanceAdherence <dbl> 0.911, 0.911, 0.911, 0.911, 0.911, 0.9…
$ meanDetectionTime <dbl> 122, 122, 122, 122, 122, 122, 122, 122…
$ avgDecolonizationTime <dbl> 387, 387, 387, 387, 387, 387, 387, 387…
$ scale2 <dbl> 23.52147, 23.52147, 23.52147, 23.52147…
$ scale1 <dbl> 3.41952, 3.41952, 3.41952, 3.41952, 3.…
$ probSurveillanceDetection <dbl> 0.8, 0.8, 0.8, 0.8, 0.8, 0.8, 0.8, 0.8…
$ burnInPeriod <dbl> 3650, 3650, 3650, 3650, 3650, 3650, 36…