source("functions.R")Methodology
Introduction
Here we provide details on our methodology for calibrating and running our forecast, as well as a description of the technologies powering the epiworld-forecasts tool.
Forecast Methodology
We use case count data published weekly by the Utah DHHS. The calibration and SIR connected model are run using the epiworldR package.
COVID-19 Cases in Utah (last 90 days)
Every week, Utah DHHS publishes COVID-19 surveillance data on their Coronavirus Dashboard which includes reported case counts for each day starting March 18, 2020. Our forecast is calibrated on case counts from the last 90 days.
# Define forecast parameters
n_days <- 90 # Calibrate model last 90 days of data
# Get COVID-19 data
covid_data <- get_covid_data(n_days)
plot_covid_data(covid_data)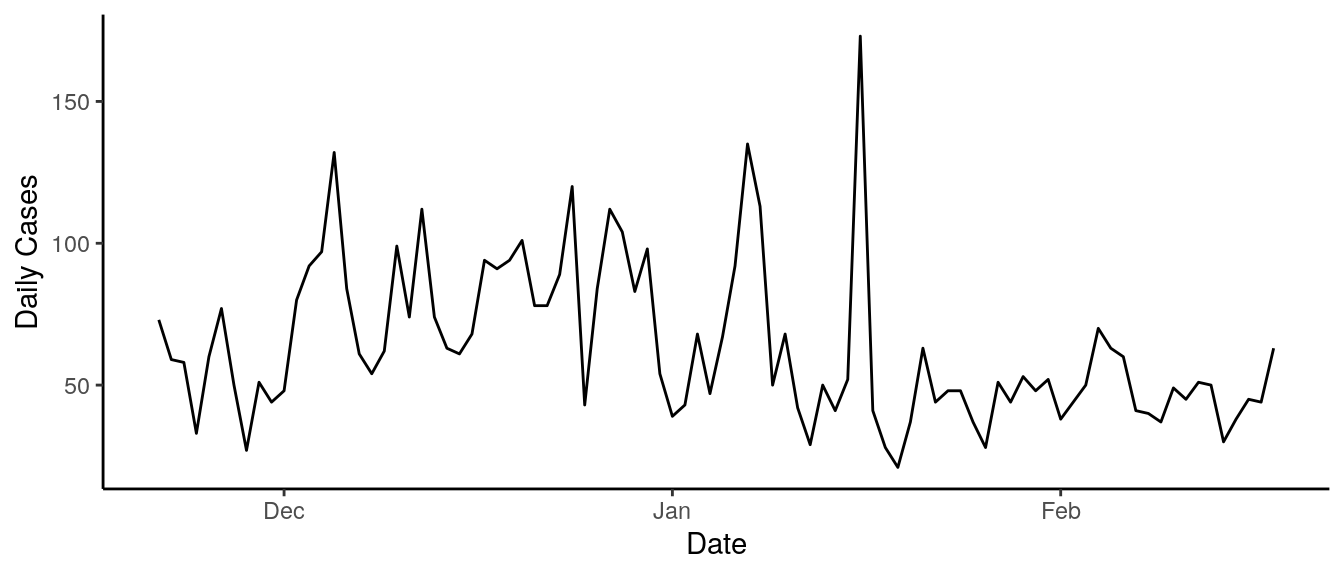
Calibrating the Forecasting Model
We use epiworldR’s implementation of Likelihood-Free Markhov Chain Monte Carlo (LFMCMC) to calibrate the SIR connected model (using ModelSIRCONN from epiworldR) by estimating the following model parameters:
- Recovery rate
- Transmission rates for each season (spring, summer, fall, winter)
- Contact rates for weekdays and weekends
The LFMCMC simulation runs for 6,000 iterations. In each iteration, it does the following:
- Proposes a new set of parameter values
- Runs the model with 10,000 agents and the proposed parameters
- Compares the output to the UDHHS data and records the result
We note that while the population of Utah over 3 million, we only need to run our model with 10,000 agents. This is because we don’t expect more than 10,000 COVID-19 cases and the model automatically scales the contact rate to account for the difference between the model population and Utah’s population.
COVID-19 Forecast
We start by preparing the model
# Compute start date for each season
seasons <- get_season_starts(covid_data$Date)
# Get observed case counts
covid_cases <- covid_data$Daily.Cases
# Define SIRCONN model parameters
model_seed <- 112 # Random seed
model_ndays <- n_days # How many days to run the model
model_n <- 10000 # model population size
# Define initial disease parameters
init_prevalence <- covid_cases[1] / model_n
init_contact_rate <- 20
init_transmission_rate <- 0.025
init_recovery_rate <- 1 / 7
# Create the SIRCONN model
covid_sirconn_model <- ModelSIRCONN(
name = "COVID-19",
n = model_n,
prevalence = init_prevalence,
contact_rate = init_contact_rate,
transmission_rate = init_transmission_rate,
recovery_rate = init_recovery_rate
)We then continue setting the model parameters. The definition of the model itself is in the functions.R script.
# Define LFMCMC parameters
lfmcmc_n_samples <- 4000 # number of LFMCMC iterations
lfmcmc_burnin <- floor(lfmcmc_n_samples / 2) # burn-in period
lfmcmc_epsilon <- 0.25
init_lfmcmc_params <- c(
1 / 7, # r_rate
0.025, # t_rate_spring
0.02, # t_rate_summer
0.03, # t_rate_fall
0.035, # t_rate_winter
20, # c_rate_weekday
2 # c_rate_weekend
)
param_names <- c(
"Recovery rate",
"Transmission rate (spring)",
"Transmission rate (summer)",
"Transmission rate (fall)",
"Transmission rate (winter)",
"Contact rate (weekday)",
"Contact rate (weekend)"
)
stats_names <- c(
"Time to peak",
"Size of peak",
"Mean (cases)",
"Standard deviation (cases)"
)With all the needed components for the LFMCMC, we can now run the calibration.
# Create the LFMCMC object
calibration_lfmcmc <- LFMCMC(covid_sirconn_model) |>
set_simulation_fun(lfmcmc_simulation_fun) |>
set_summary_fun(lfmcmc_summary_fun) |>
set_proposal_fun(lfmcmc_proposal_fun) |>
set_kernel_fun(lfmcmc_kernel_fun) |>
set_observed_data(covid_cases)
# Run LFMCMC calibration
run_lfmcmc(
lfmcmc = calibration_lfmcmc,
params_init = init_lfmcmc_params,
n_samples = lfmcmc_n_samples,
epsilon = lfmcmc_epsilon,
seed = model_seed
)_________________________________________________________________________
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| done.set_params_names(calibration_lfmcmc, param_names)
set_stats_names(calibration_lfmcmc, stats_names)When the simulation is finished, we use a burn-in period of n = 2,000 (50% of simulation iterations). The epiworldR results printout (below) shows the mean parameter/statistic value, the 95% credible interval (in [ ]), and the initial/observed value (in ( )).
print(calibration_lfmcmc, burnin = lfmcmc_burnin)___________________________________________
LIKELIHOOD-FREE MARKOV CHAIN MONTE CARLO
N Samples (total) : 4000
N Samples (after burn-in period) : 2000
Elapsed t : 3.00m
Parameters:
-Recovery rate : 0.33 [ 0.26, 0.39] (initial : 0.14)
-Transmission rate (spring) : 0.01 [ 0.01, 0.02] (initial : 0.03)
-Transmission rate (summer) : 0.03 [ 0.02, 0.05] (initial : 0.02)
-Transmission rate (fall) : 0.09 [ 0.06, 0.13] (initial : 0.03)
-Transmission rate (winter) : 0.03 [ 0.03, 0.04] (initial : 0.04)
-Contact rate (weekday) : 19.46 [ 18.68, 20.19] (initial : 20.00)
-Contact rate (weekend) : 0.64 [ 0.25, 0.95] (initial : 2.00)
Statistics:
-Time to peak : 21.12 [ 1.00, 22.00] (Observed: 19.00)
-Size of peak : 137.33 [ 60.00, 198.00] (Observed: 162.00)
-Mean (cases) : 15.04 [ 7.21, 26.56] (Observed: 38.78)
-Standard deviation (cases) : 28.38 [ 13.94, 41.10] (Observed: 18.70)
___________________________________________Looking into the trace of the MCMC chain
accepted <- get_all_accepted_params(calibration_lfmcmc)
accepted <- lapply(seq_len(ncol(accepted)), \(i) {
data.frame(
iteration = 1:nrow(accepted),
name = param_names[i],
value = accepted[, i]
)
}) |> do.call(what = "rbind")
ggplot(accepted, aes(x = iteration, y = value, color = name)) +
geom_line() +
labs(title = "LFMCMC Trace Plot",
x = "Iteration",
y = "Parameter value") +
facet_wrap(~name, scales = "free_y") +
theme_minimal()
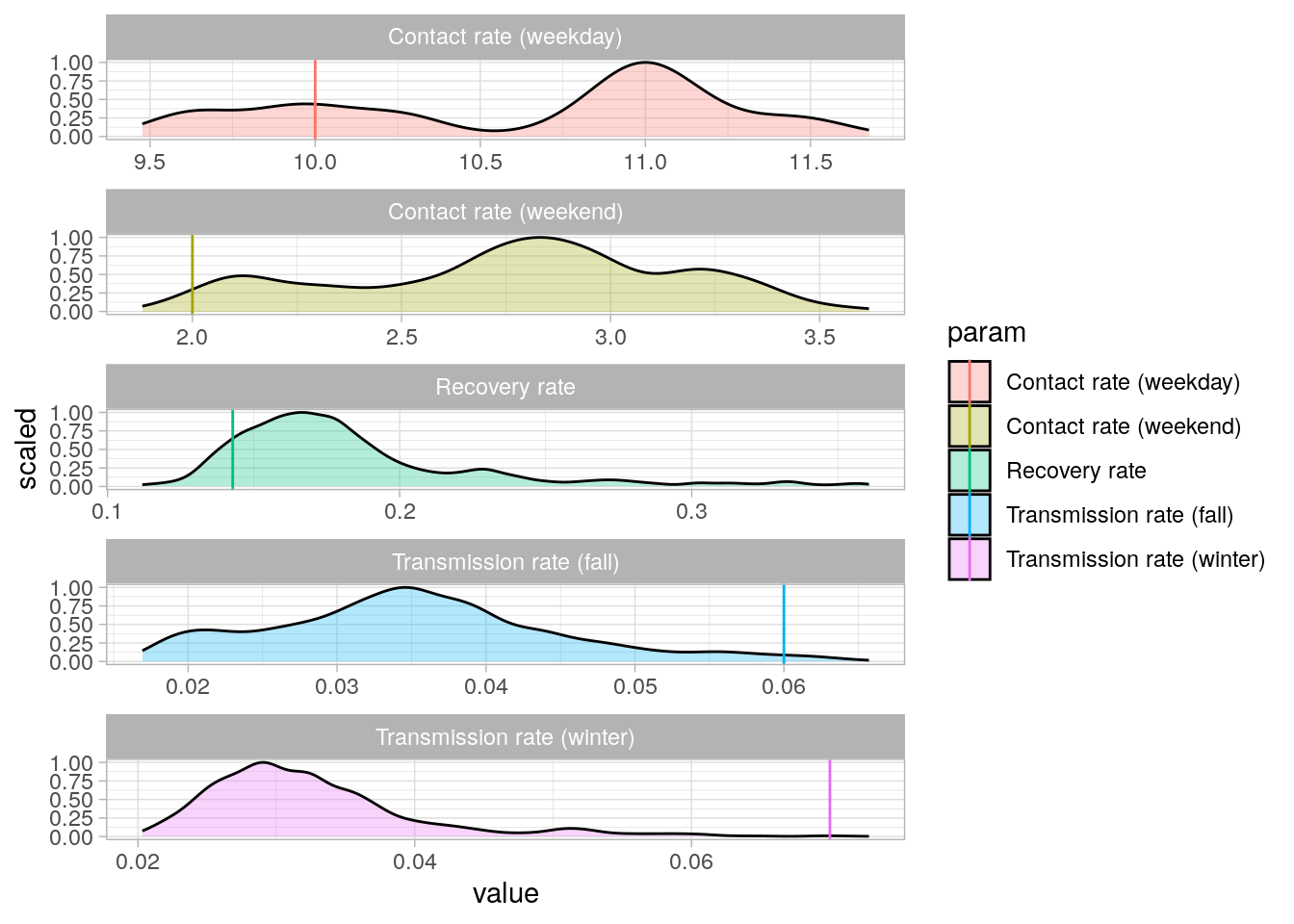
Here is the posterior distribution of the LFMCMC samples with vertical lines representing the initial parameter values.
plot_lfmcmc_post_dist(
calibration_lfmcmc,
init_lfmcmc_params,
param_names,
seasons
)
We can now run the forecast.
# Create a new SIR CONN model
# - Compute prevalence based on most recent day
forecast_prevalence <- covid_cases[90] / model_n
# - Init the model
covid_sirconn_model <- ModelSIRCONN(
name = "COVID-19",
n = model_n,
prevalence = forecast_prevalence,
contact_rate = init_contact_rate,
transmission_rate = init_transmission_rate,
recovery_rate = init_recovery_rate
)
# Run the simulation for each set of params in the sample
# - Select sample of accepted params from LFMCMC
forecast_sample_n <- 200 # Sample size
params_sample <- get_params_sample(calibration_lfmcmc,
lfmcmc_n_samples,
lfmcmc_burnin,
forecast_sample_n)
# - Set forecast length
model_ndays <- 14
# - Run simulation function for each set of params from the sample
forecast_dist <- apply(params_sample, 1, lfmcmc_simulation_fun)Our model prevalence is set according to the reported case counts of the most recent day of the UDHHS data. We then take a sample of n = 200 from the LFMCMC accepted parameters (after the burn-in period) and run the SIR connected model with the new prevalence for each set of parameters. Each simulation is for two weeks, giving us a 14-day forecast of COVID-19 in Utah. The forecast mean is shown below along with the 50% and 95% confidence intervals. The actual case counts are plotted in black, while the forecast is plotted in blue.
plot_forecast(forecast_dist, covid_data)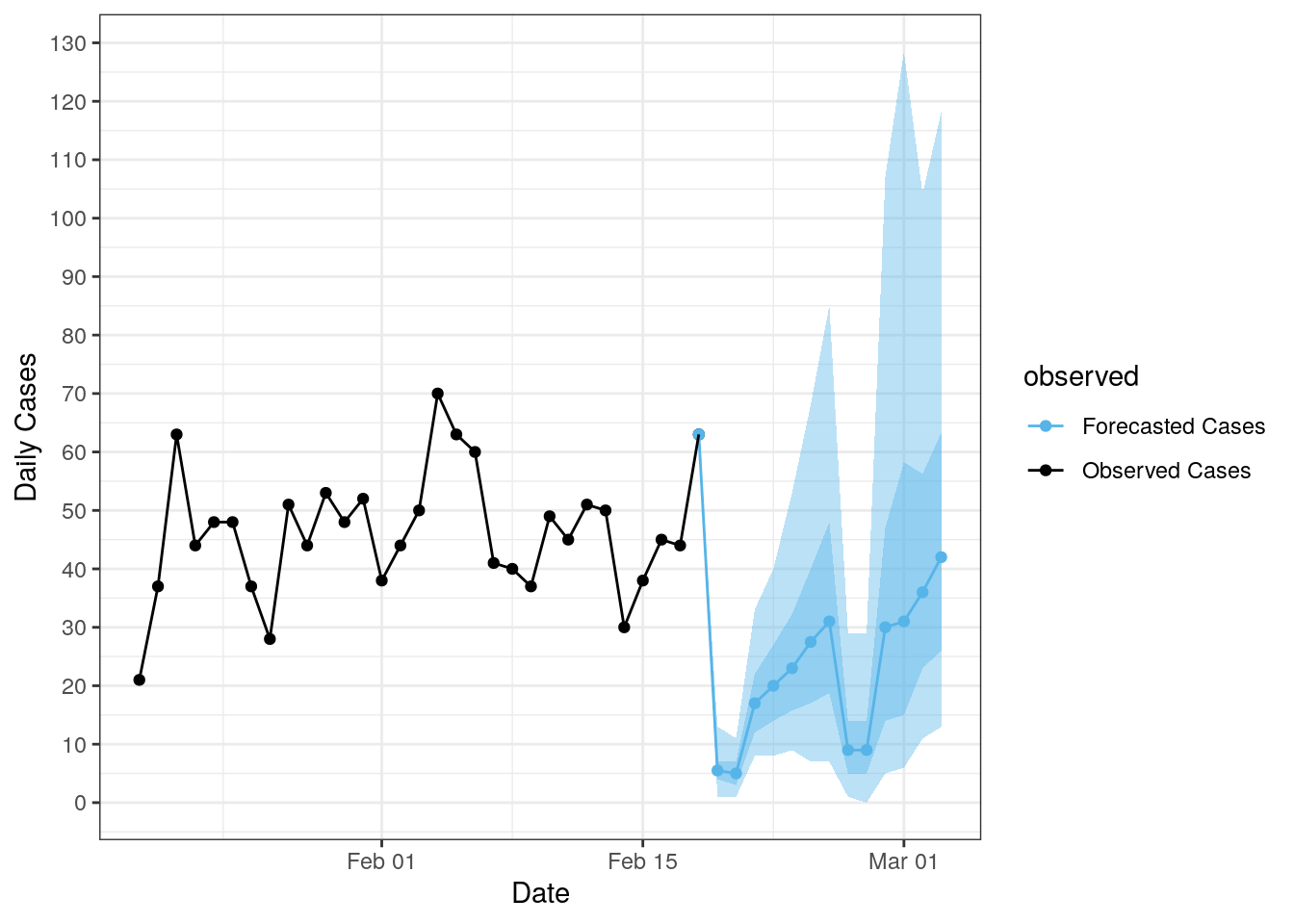
saveRDS(
list(
forecast_dist = forecast_dist,
covid_data = covid_data
),
sprintf(
"forecast_data_%s.rds",
format(Sys.time(), "%Y-%m-%d")
)
)Technologies
epiworld-forecasts is built with the following technologies:
- epiworldR: Fast agent-based modeling R package for disease simulations
- Docker: The forecast runs inside a Docker container which has all the needed packages. This container is built in a separate workflow and pushed to the GitHub Container Registry.
- GitHub Actions: The forecast runs on a schedule through GitHub Actions
- Quarto: Generates the HTML report that is published to GitHub Pages
- GitHub: Version control, hosting the source code repository and website