library(readr) # tidyverse's
library(data.table)
library(microbenchmark)
# Generating a large dataset of random numbers
set.seed(1231)
x <- runif(5e4 * 10) |> matrix(ncol = 10) |> data.frame()
# Creating tempfiles
temp_dt <- tempfile(fileext = ".csv")
temp_tv <- tempfile(fileext = ".csv")
temp_r <- tempfile(fileext = ".csv")The data.table R package
For most cases, the most proper data wrangling tool in R is the dplyr package. Nonetheless, when dealing with large amounts of data, the data.table package is the fastest alternative available for doing data processing within R (see the benchmarks).
Reading and Writing Data
Reading and writing operations with data.table’s fread and fwrite are highly optimized. Here is a benchmark we can do on our own:
bm <- microbenchmark(
readr = write_csv(x, temp_tv, num_threads = 1L, progress = FALSE),
data.table = fwrite(
x, temp_dt, verbose = FALSE, nThread = 1L,
showProgress = FALSE),
base = write.csv(x, temp_r),
times = 20
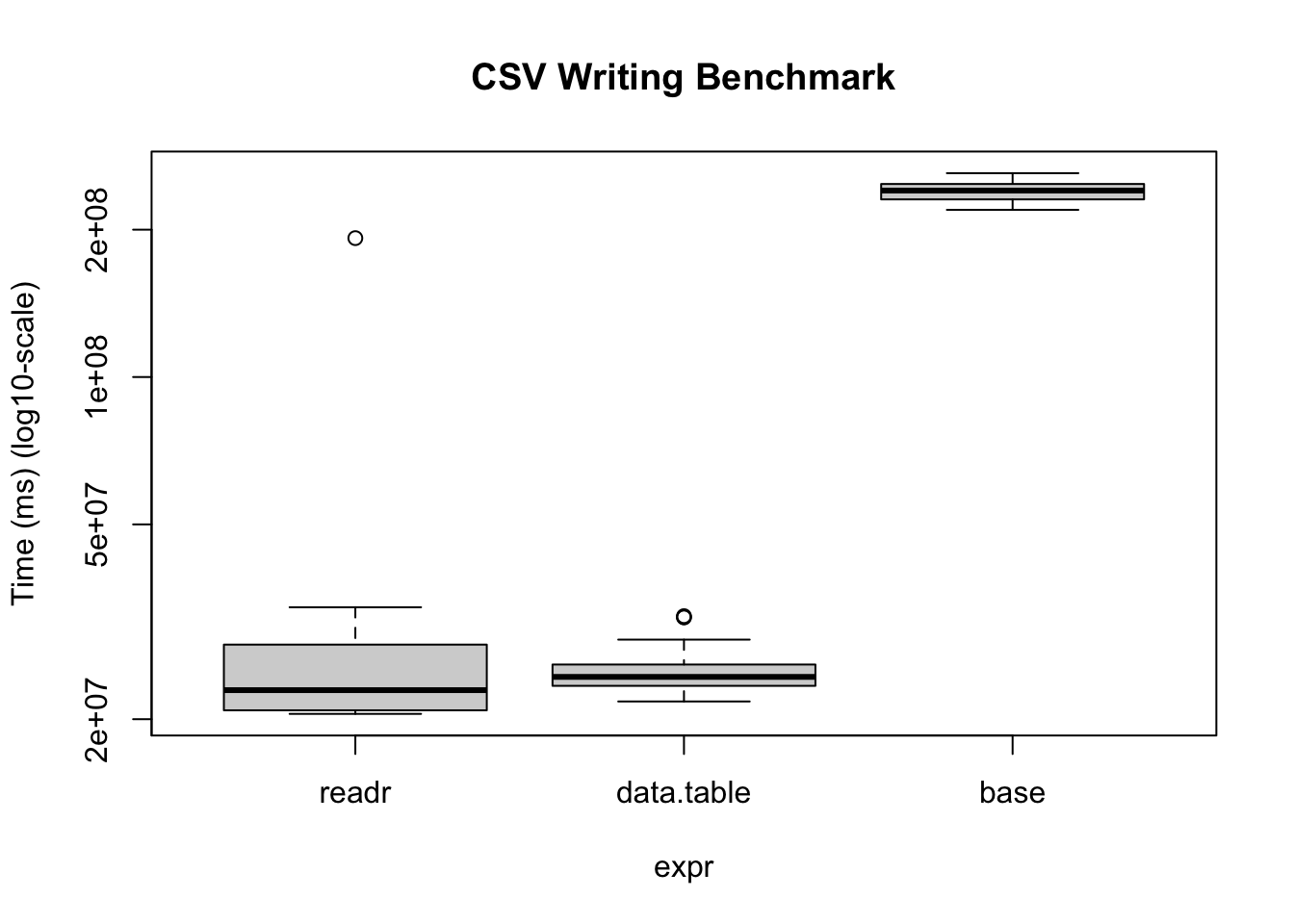
)Warning in microbenchmark(readr = write_csv(x, temp_tv, num_threads = 1L, :
less accurate nanosecond times to avoid potential integer overflowsbmUnit: milliseconds
expr min lq mean median uq max neval
readr 20.50656 20.85293 32.74184 22.92738 28.40509 192.2000 20
data.table 21.73935 23.39716 25.49071 24.40361 25.86483 32.4754 20
base 219.56336 230.62471 239.80889 240.37378 247.95769 260.8928 20# We can also visualize it
bm |>
plot(
log = "y",
ylab = "Time (ms) (log10-scale)",
main = "CSV Writing Benchmark"
)
The same thing applies when reading data
# Writing the data
fwrite(x, temp_r, verbose = FALSE, nThread = 1L, showProgress = FALSE)
# Benchmarking
bm <- microbenchmark(
readr = read_csv(
temp_r, progress = FALSE, num_threads = 1L,
show_col_types = FALSE
),
data.table = fread(
temp_r, verbose = FALSE, nThread = 1L,
showProgress = FALSE
),
base = read.csv(temp_r),
times = 10
)Warning in microbenchmark(readr = read_csv(temp_r, progress = FALSE,
num_threads = 1L, : less accurate nanosecond times to avoid potential integer
overflowsbmUnit: milliseconds
expr min lq mean median uq max neval
readr 40.088898 40.28516 74.29501 41.79661 42.52569 370.04095 10
data.table 9.947297 10.24852 11.92535 10.63159 11.27336 21.29651 10
base 148.565591 176.56457 204.73412 199.02650 235.19945 247.29724 10bm |>
plot(
log = "y",
ylab = "Time (ms) (log10-scale)",
main = "CSV Reading Benchmark"
)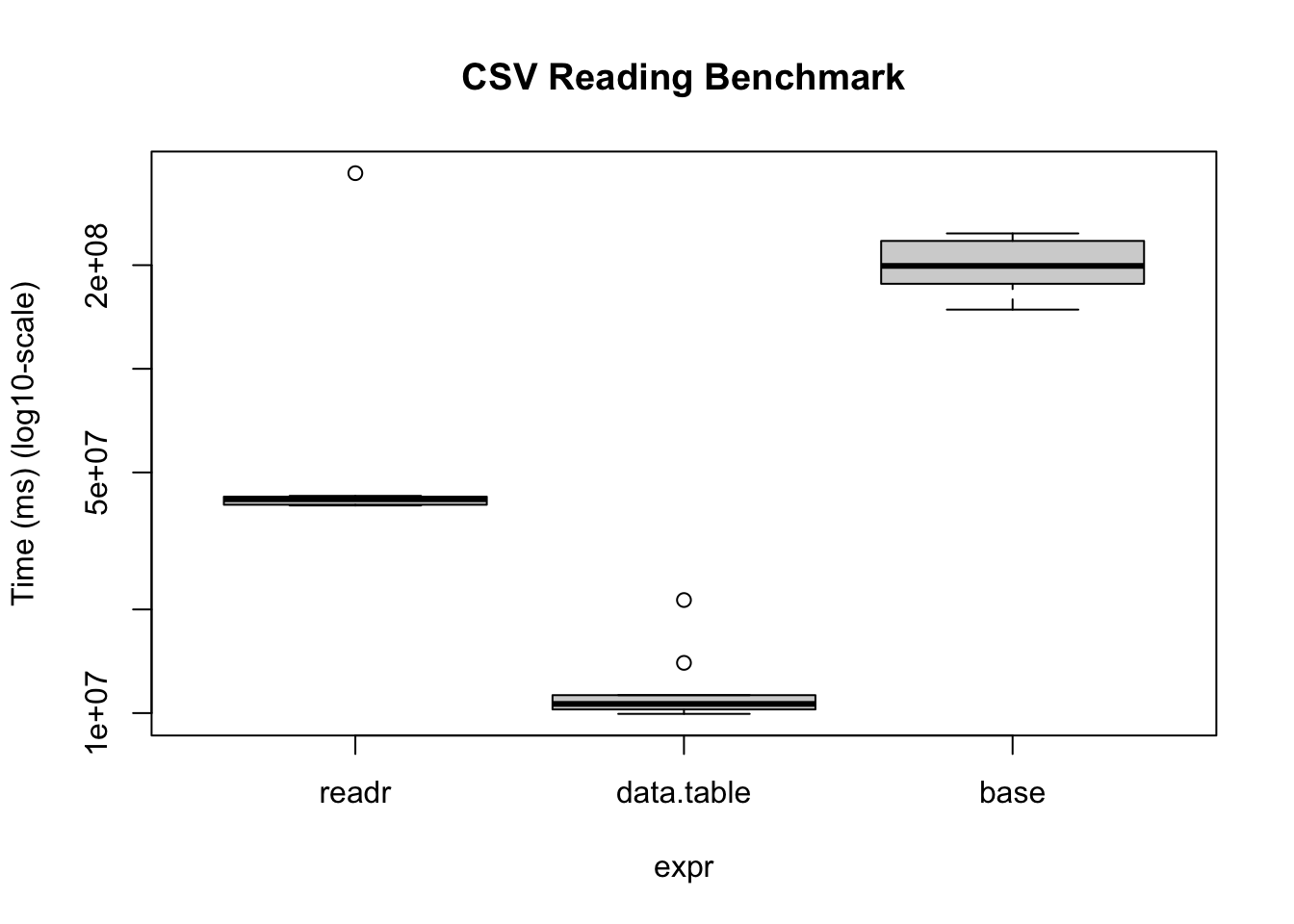
Under the hood, the readr package uses the vroom package. Nonetheless, there are some operations (dealing with character data mostly), where the vroom package shines. Regardless, data.table is a perfect alternative as it goes beyond just reading/writing data.
Example data manipulation
data.table is also the fastest for data manipulation. Here are a couple of examples aggregating data with data table vs dplyr
input <- if (file.exists("flights14.csv")) {
"flights14.csv"
} else {
"https://raw.githubusercontent.com/Rdatatable/data.table/master/vignettes/flights14.csv"
}
# Reading the data
flights_dt <- fread(input)
flights_tb <- read_csv(input, show_col_types = FALSE)library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:data.table':
between, first, lastThe following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, union# To avoid some messaging from the function
options(dplyr.summarise.inform = FALSE)
microbenchmark(
data.table = flights_dt[, .N, by = .(origin, dest)],
dplyr = flights_tb |>
group_by(origin, dest) |>
summarise(n = n())
)Unit: milliseconds
expr min lq mean median uq max neval
data.table 1.788051 2.121955 2.789121 2.367279 2.963931 5.913922 100
dplyr 5.255011 5.544102 6.682056 5.832742 6.371748 37.637713 100